mrna <- read.table("data/data_example.txt", header=T, sep="\t", dec=".")
# Convert to matrix format
mrna <- as.matrix(mrna)Practical applications of learned concepts in R
Example
TBD
Please run the code provided to replicate some of the analyses. Make sure you can explain what all the analysis steps do and that you understand all the results.
In addition, there are some extra tasks (Task 1), where no R code is provided. Please do these tasks when you have time available at the end of the lab.
Load the data
Read the data, and convert to matrix format.
Print the data
mrna[1:4, 1:4] OSL2R.3002T4 OSL2R.3005T1 OSL2R.3013T1 OSL2R.3030T2
ACACA 1.60034 -0.49087 -0.26553 -0.27857
ANXA1 -2.42501 -0.05416 -0.46478 -2.18393
AR 0.39615 -0.43348 -0.10232 0.58299
BAK1 0.78627 0.39897 0.22598 -1.31202Visualize the overall distribution of expression levels by histogram
hist(mrna, nclass=40, xlim=c(-5,5), col="lightblue")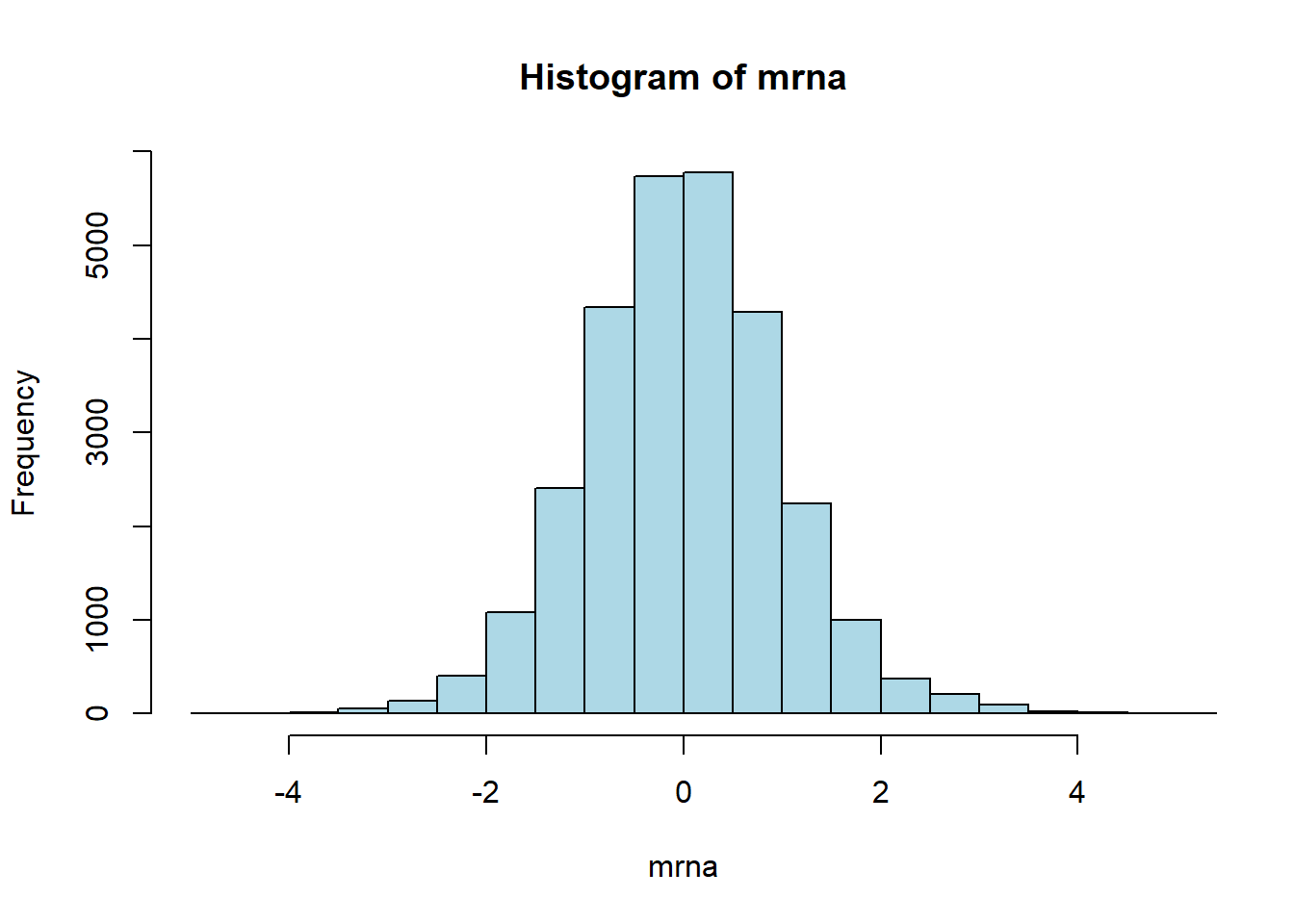
Task 1
This is a callout-note, and it can be quite useful for exercises. You can find more about callout here.
Example: Extend the above analysis to cover all genes.